Lab members

Natalia Sharova
Head of the Lab, Doctor of Biological Sciences
Main research interests: proteasome mechanisms of mammalian organ and system development, the role of proteasomes in immune system functioning and cancer growth.

Victor Mikhailov
Chief researcher, Profeesor, Doctor of Biological Sciences
Main research interests: proteome regulation in lepidopteran insect Spodoptera frugiperda Sf9 cells at normal conditions and under infection with baculovirus.

Liudmila Zakharova
Chief researcher, Professor, Doctor of Biological Sciences
Main research interests: cellular and molecular mechanisms of neuroendocrine-immune interactions in ontogenesis.

Yegor Vassetzky
Leading researcher, Doctor of Biological Sciences
Main research interests: concern the role of the nuclear organization and epigenetic regulation in cancer and neuromuscular diseases.

Yulia Lyupina
Chief researcher, Doctor of Biological Sciences
Main research interests: sponge regeneration, iron metabolism proteins, ubiquitin proteasome system.

Wladyslaw Krajewski
Leading researcher, Doctor of Biological Sciences
Main research interests: the structure-functional relationship in chromatin, and particularly, the (cross) effects of histone modifications and enzymatic remodeling on chromatin functional activity, structure, and dynamics.

Pavel Erokhov
Leading researcher, PhD
Main research interests: biochemistry of redox and free radical processess, cell biology.

Tatiana Astakhova
Senior researcher, PhD
Main research interests: protein metabolism and proteasomes in cancer, molecular mechanisms of the development of tolerance to tumors.

Oksana Kravchuk
Senior researcher, PhD
Main research interests: molecular genetics of drosophila, regulation of gene expression.

Marina Izvolskaia
Senior researcher, PhD
Main research interests: effects of the intrauterine inflammation on the reproductive axis development and functioning in mouse and rat offspring.

Yaroslava Karpova
Researcher, PhD
Main research interests: proteasomes in the development of immune system and cancer cells.

Kim Adameyko
Junior researcher
Main research interests: bioinformatics, transcriptomics, chromatin, iron metabolism, invertebrates.

Vasilina Ignatyuk
Researcher, PhD
Main research interests: long-term effects of inflammation on the development of hypothalamic-pituitary system.

Alexander Finoshin
Researcher, PhD
Main research interests: cell plasticity, proteomics, cell homeostasis proteomics, regeneration, metalloproteinases, simbiotic bacteria.

Victoria Sharova
Junior researcher
Main research interests: role of cytokines in GnRH development under normal and pathological conditions.

Svetlana Abaturova (Dmitrieva)
Research assistant, PhD
Main research interests: proteasomes, DNA-polymerases, reparative DNA-polymerases in the embryogenesis of teleost fish.

Vera Gorelova
Research assistant
Main research interests:

Anna Karpuhina
Junior researcher, PhD student
Main research interests: molecular mechanisms of cancerogenesis, muscle dystrophies, transcriptomics, bistable systems.
Research fields
Proteasomes in early ontogenesis and tumor development
One of the main research directions in the Laboratory of Developmental Biochemistry is the study of the proteasome mechanisms of mammalian organ and system development. Proteasomes, which are multisubunit proteases, are the main players in protein hydrolysis in a cell. Proteasomes control the cellular proteome and regulate different processes by protein degradation and production of biologically active polypeptides and peptides. The multiple functions of proteasomes result from the multiplicity of their forms, different in the structure and specificity of the hydrolysis of protein substrates. According to the set of proteolytic subunits, the proteasome pool in mammalian cells may be divided into constitutive (or “housekeeping”) proteasomes and immune ones. The immune proteasomes integrate immune proteolytic subunits LMP2(β1i), MECL1(β2i), and LMP7(β5i) instead of constitutive subunits β1, β2, and β5, respectively, in the process of new proteasome assembling. Interestingly, the immune subunits can be embedded in proteasomes either together or in various combinations with the constitutive subunits, generating several subtypes of immune proteasomes.
Proteasomes in the development of the thymus, spleen, and liver were investigated in our Laboratory (Sharova et al., 2009; Melnikova et al., 2010). Understanding the proteasome mechanisms of the early ontogenesis processes is important for the detection of mechanisms underlying the development of different pathologies.
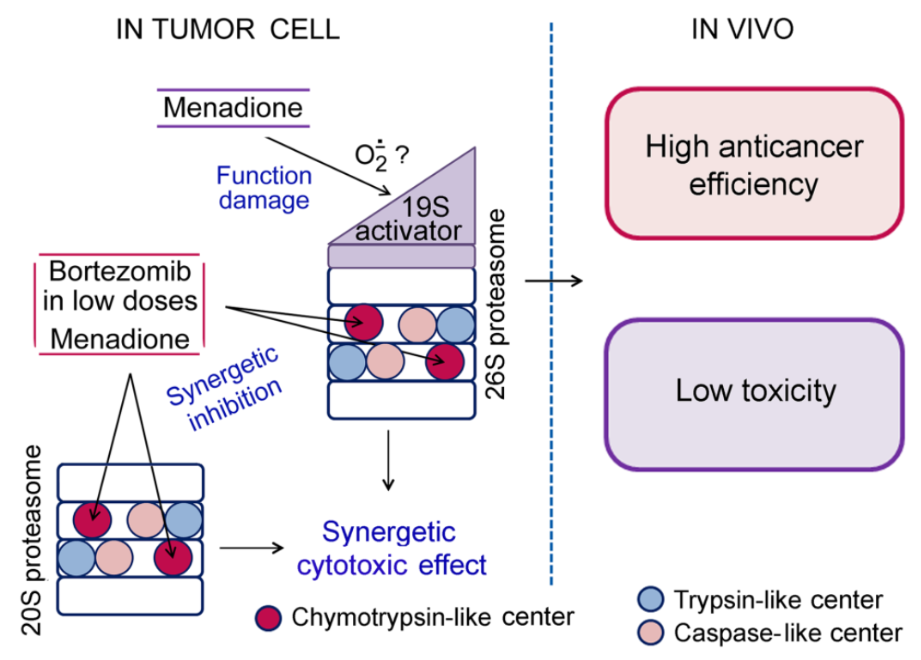
We have recently discovered an important connection of the proteasome subtype containing the LMP2 (but not LMP7) subunit with the development of the immune tolerance to the “foreign” (Astakhova et al., 2019). This fact has allowed us to look at tumor growth mechanisms in a new way (Erokhov et al., 2021). Besides, novel targets for antitumor therapy and markers for tumor diagnostics are being investigated. Thus, we have developed a new composition of bortezomib and menadione sodium bisulfite (MSB) with a combined action on the proteasomes of tumor cells: MSB indirectly suppressed the 26S proteasome function, and MSB together with bortezomib inhibited the chymotrypsin-like activity of the total proteasome pool in a synergetic manner. These influences led to a synergetic cytotoxic effect (Astakhova et al., 2018).
Recent publications
- Sharova N.P., Zakharova L.A., Astakhova T.M., Karpova Ya.D., Melnikova V.I., Dmitrieva S.B., Lyupina Yu.V., Erokhov P.A. New approach to study of T cellular immunity development: Parallel investigation of lymphoid organ formation and changes in immune proteasome amount in rat early ontogenesis // Cell. Immunol. 2009. V. 256. P. 47–55.
- Melnikova V., Sharova N., Maslova E., Voronova S., Zakharova L. Ontogenesis of Rat Immune System: proteasome expression in different cell populations of the developing thymus // Cell. Immunol. 2010. V. 266. P. 83–89.
- Zakharova L.A., Khegai I.I., Sharova N.P., Melnikova V.I., Karpova Y.D., Astakhova T.M., Popova N.A., Ivanova L.N. Pattern of MHC class I and immune proteasome expression in Walker 256 tumor during growth and regression in Brattleboro rats with the hereditary defect of arginine-vasopressin synthesis // Cell. Immunol. 2011. V. 271. P. 385-391.
- Sharova N.P., Astakhova T.M., Karpova Ya.D., Lyupina Yu.V., Alekhin A.I., Goncharov N.G., Sumedi I.R., Cherner V.A., Rodoman G.V., Kuznetsov N.A., Erokhov P.A.. Changes in proteasome pool in human papillary thyroid carcinoma development // Cent. Eur. J. Biol. 2011. V. 6, No 4, P. 486-496.
- Shashova E.E., Lyupina Yu.V., Glushchenko S.A., Slonimskaya E.M., Savenkova O.V., Kulikov A.M., Gornostaev N.G., Kondakova I.V., Sharova N.P. Proteasome functioning in breast cancer: connection with clinical-pathological factors // Plos One. Published: October 17, 2014, DOI: 10.1371/journal.pone.0109933.
- Karpova Ya. D., Bozhok G. A., Alabedal’karim N. M., Lyupina Yu. V., Astakhova T. M., Legach E. I., and Sharova N. P..Proteasomes and Transplantology: Current state of the problem and the search for promising trends // Biology Bulletin. 2017. V. 44. No. 3. P. 237–244.
- Erokhov P.A., Lyupina Yu.V., Radchenko A.S., Kolacheva A.A., Nikishina Yu.O., Sharova N.P. Detection of active proteasome structures in brain extracts: Proteasome features of August rat brain with violations in monoamine metabolism // Oncotarget. 2017; 8:70941-70957. https://doi.org/10.18632/oncotarget.20208.
- Astakhova T.M., Morozov A.V., Erokhov P.A., Mikhailovskaya M.I., Akopov S.B., Chupikova N.I., Safarov R.R, Sharova N.P. Combined effect of bortezomib and menadione sodium bisulfite on proteasomes of tumor cells: the dramatic decrease of bortezomib toxicity in a preclinical trial // Cancers. 2018. V. 10. Issue 10. pii: E351. doi: 10.3390/cancers10100351.
- Astakhova T.M., Bozhok G.A., Alabedal’karim N.M., Karpova Ya.D., Lyupina Yu.V., Ushakova E.M., Legach E.I., Bondarenko T.P., Sharova N.P. Proteasome expression in ovarian heterotopic allografts of Wistar and August rats under induction of donor specific tolerance // Russian Journal of Developmental Biology. 2019. V. 50. No. 5. P. 261-267. DOI: 10.1134/S1062360419050023
- Kondakova I.V., Shashova E.E., Sidenko E.A., Astakhova T.M., Zakharova L.A., Sharova N.P. Estrogen receptors and ubiquitin proteasome system: mutual regulation // Biomolecules. 2020. V. 10. № 4. pii: E500. DOI: 10.3390/biom10040500.
- Erokhov P.A., Kulikov A.M., Karpova Y.D., Rodoman G.V., Sumedi I.R., Goncharov A.L., Razbirin D.V., Gorelova V.S., Sharova N.P., and Astakhova T.M. Proteasomes in patient rectal cancer and different intestine locations: Where does proteasome pool change?//Cancers. 2021. V. 13. P. 1108. https://doi.org/10.3390/cancers13051108
Сellular and molecular mechanisms of neuroendocrine-immune interactions in ontogenesis
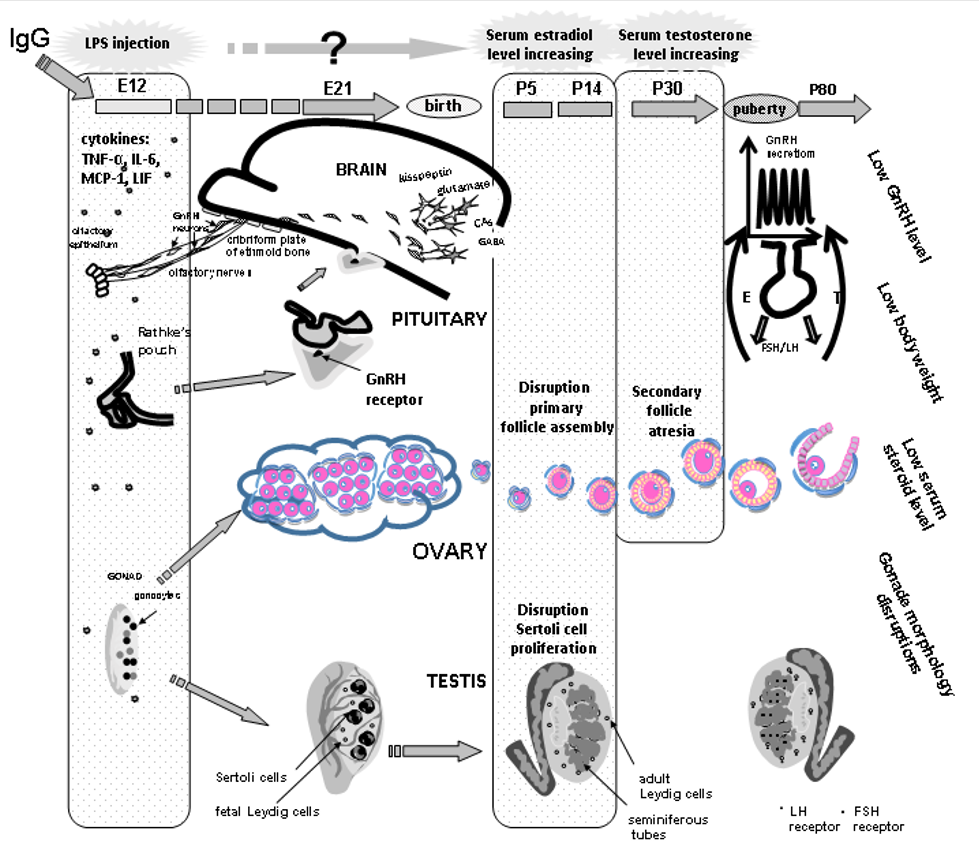
Fig. 2. The mechanisms underlying the developmental origins of female and male postnatal sexual abnormalities after prenatal exposure to lipopolysaccharide (LPS). GnRH – gonadotropin-releasing hormone, E12… – days of the embryonic development, P5… – days of the postnatal development, LH – luteinizing hormone, FSH – follicle stimulating hormone, E – estradiol, IgG – immunoglobulin G, cytokines: IL-6 interleukin, LIF leukemia inhibitory factor, MCP-1 monocyte chemotactic protein, TNFα tumor necrosis factor.
A long-term interest of Professor Liudmila A. Zakharova and her colleagues focuses on the effects of the intrauterine inflammation induced by lipopolysaccharide (LPS, Escherichia coli), a major component of the outer membrane in gram-negative bacteria, and the possible mechanisms underlying the LPS effect on the hypothalamic–pituitary–gonadal axis development and its functioning in adult offspring. LPS is one of the most potent natural inducers of inflammation coupled with an increased synthesis of pro- and anti-inflammatory cytokines in both maternal and fetal organisms. LPS-induced increases in fetal levels of proinflammatory cytokines can affect brain development and have long-term effects on behavior and neuroendocrine functions in offspring. According to our data, an LPS-induced increase in the levels of interleukin (IL)-6, tumor necrosis factor α (TNFα), leukemia inhibitory factor (LIF), and monocyte chemotactic protein (MCP-1) in maternal and fetal mice during early pregnancy disturbs the development and functioning of gonadotropin-releasing hormone (GnRH) production and reproductive systems. Prenatal LPS treatment delays the intranasal migration of GnRH neurons into the brain and affects testicular and ovarian development in rats, which can be attributed to the high levels of proinflammatory cytokines in the fetus, and sex steroids in the prepubertal period. Antagonists of receptors for testosterone (flutamide) and estradiol (fulvestrant) administered in the period of elevated hormones to females and males, respectively, normalize sexual maturation in adult offspring, thus almost completely restoring their sexual gland structure, sex hormone levels, and reproductive capacity (Figure). In addition, IgG is able to modulate inflammation processes in early ontogenesis. Thus, reproductive abnormalities in adults should be controlled and prevented in both prenatal and infantile periods.
Recent publications
- Zakharova L., Sharova V., Izvolskaia M. (2021) Mechanisms of reciprocal regulation of gonadotropin-releasing hormone (GnRH)-producing and immune systems: the role of GnRH, cytokines and their receptors in early ontogenesis in normal and pathological conditions. Int. J. Mol. Sci., 22, 114. https://doi.org/10.3390/ijms22010114.
- Izvolskaia M.S., Ignatiuk V.M., Ismailova A.H., Sharova V.S., Zakharova L.A. (2021) IgG immune modulation in male mice with reproductive failure after prenatal inflammation. Reproduction. https://doi.org/10.1530/REP-20-0386.
- Izvolskaia M.S., Sharova V.S., Zakharova L.A. (2020) Perinatal Inflammation Reprograms Neuroendocrine, Immune, and Reproductive Functions: Profile of Cytokine Biomarkers. Inflammation. 43, 1175–1183. doi: 10.1007/s10753-020-01220-1.
- Izvolskaia M.S., Sharova V.S., Ignatiuk V.M., Voronova S.N., Zakharova L.A. (2019) Abolition of prenatal lipopolysaccharide-induced reproductive disorders in rat male offspring by fulvestrant. Andrologia. 51, e13204. http://doi.10.1111/and.13204.
- Ignatiuk V.M., Izvolskaya M.S., Sharova V.S., Voronova S.N., Zakharova L.A. (2019) Disruptions in the reproductive system of female rats after prenatal lipopolysaccharide-induced immunological stress: role of sex steroids. Stress. 22, 133–141. doi: 10.1080/10253890.2018.1508440.
- Melnikova V.I., Lifantseva N.V., Voronova S.N., Zakharova L.A. (2019) Gonadotropin-Releasing Hormone in regulation of thymic development in rats: Profile of thymic cytokines. Int. J. Mol. Sci. 20(16), 4033. doi 10.3390/ijms20164033.
- Izvolskaia M.S., Tillet Y., Sharova V.S., Voronova S.N., Zakharova L.A. (2016) Disruptions in the hypothalamic-pituitary-gonadal axis in rat offspring following prenatal maternal exposure to lipopolysaccharide. Stress, 19, 198–205. doi: 10.3109/10253890.2016.1149695.
- Sharova V.S., Izvolskaia M.S., Zakharova L.A. (2015) Lipopolysaccharide-induced maternal inflammation affects the gonadotropin-releasing hormone neuron development in fetal mice. Neuroimmunomodulation. 22, 222–232. doi: 10.1159/000365482.
- Zakharova L. (2015) Perinatal stress in brain programming and pathogenesis of psychoneurological disorders. Biol. Bull. 42, 12–20.
- Zakharova L.A., Izvolskaia M.S. (2012) Interactions between the reproductive and immune systems during ontogenesis: The role of GnRH, sex steroids and immunomediators. In Sex steroids; Kahn, S.M., Ed.; InTech: Rijeka, Croatia; London, UK, 2012, pp. 341–370.
The structure-function relationship in chromatin
A long-term interest of Wladyslaw Krajewski is the structure-functional relationship in chromatin and, particularly, the (cross) effects of histone modifications and enzymatic remodeling on chromatin functional activity, structure, and dynamics. The research was supported by several international research fellowships and awards, which were held in recognized scientific centers. Our work in the past mostly focused on the in vitro biochemical characterization of the nucleosome structure and dynamics (and its relation to chromatin functions) and on the mechanisms of mono- and polynucleosome remodeling by yeast ATP-dependent enzymes Isw1/Isw2 and Swi-Snf (see Recent publications). Besides providing data of “basic” scientific importance, our studies also suggested some potential practical approaches (such as the strategy for the “hyperactivation” of gene promoters by short oligonucleotide insertions capable of forming “non-canonical” DNA structures, as well as some helpful experimental tools and techniques (Krajewski WA 2015/2016 Analytical Biochemistry).
Recent studies focus on the “direct” effects of bulky histone modifications (ubiquitylation and sumoylation) in the nucleosome structure and dynamics in a defined in vitro system of pure components. We have shown that homogeneous semi-synthetic ubiquitylation of both nucleosomal histones H2B or (to a lesser extent) of only one of them at lysines K34 or 120 could dramatically enhance intranucleosome dynamics and promote selective eviction of (only) one nucleosome dimer (Nucleic Acids Res 2018). Consistently with nucleosome stability-effects of H2B-ubiquitylation, positive and negative DNA stresses differently affected unmodified H2BK34ub and H2BK120ub nucleosomes (J. Mol. Biol. 2018). These results highlight a novel mode of action by which histone posttranslational modifications (PTMs) regulate chromatin structure and functionality, and which is distinct from the generally accepted roles of histone modifications in the recruitment of downstream effector and in histone charge shielding (Trends Cell Biol 2019; BioEssays 42, 1900136 (commented in the Bioessays ‘Idea to Watch’ article by Jeffrey McKnight, https://doi.org/10.1002/bies.201900217). Thus, the “histone code” concept could be extended to include the PTM-programmable nucleosome conformational states that directly impact chromatin-based processes.
Recent publications
- Krajewski W.A. (2020) The intrinsic stability of H2B-ubiquitylated nucleosomes and their in vitro assembly/ disassembly by histone chaperone NAP1. Biochim Biophys Acta - Gen Subj. 1864, 1-9.
- Krajewski W.A. (2020) ‘Direct’ and ‘indirect’ effects of histone modifications: Modulation of sterical bulk as a novel source of functionality. BioEssays 42, 1900136.
- Krajewski W.A. (2019) Ubiquitylation: How nucleosomes use histones to evict histones. Trends in Cell Biology 29, 689-694
- Krajewski W.A., Vassiliev O.L. (2019) Analysis of histone ubiquitylation by MSL1/MSL2 proteins in vitro. Archives of Biochemistry and Biophysics 666, 22-30
- Krajewski W.A. (2018) Effects of DNA superhelical stress on stability of H2B-ubiquitylated nucleosomes. Journal of Molecular Biology, 430, 5002–5014
- Krajewski W.A., Li, J., and Dou, Y. (2018) Effects of histone H2B ubiquitylation on the nucleosome structure and dynamics. Nucleic Acids Research 46, 7631-7642.
- Krajewski W.A. (2016) On the role of internucleosomal interactions and intrinsic nucleosome dynamics in chromatin function. Biochemistry and Biophysics Reports 5, 492–501.
- Krajewski W.A. (2016) A composite agarose-polyacrylamide matrix as 2D hard support for solid-phase protein assays. Analytical Biochemistry 497, 57-59.
- Krajewski W.A. (2016) Mobilization of hyperacetylated mononucleosomes by purified yeast ISW2 in vitro, Archives of Biochemistry and Biophysics 591, 1-6
- Krajewski W.A. (2015) A simple and cost-effective solid-phase protein nano-assay using polyacrylamide-coated glass plates. Analytical Biochemistry 470, 78–83.
- Krajewski W.A. (2014) Yeast Isw1a and Isw1b exhibit similar nucleosome mobilization capacities for mononucleosomes, but differently mobilize dinucleosome templates. Archives of Biochemistry and Biophysics 546, 72-80
- Krajewski W.A. (2014) Isw1a does not have strict limitations on the length of extranucleosomal DNAs for mobilization of nucleosomes assembled with HeLa cell histones. J. Biomol. Structure & Dynamics 32, 523-531.
- Krajewski, W.A. (2013) Comparison of the Isw1a, Isw1b, and Isw2 nucleosome disrupting activities. Biochemistry 52, 6940-6949
- Krajewski W.A., Vassiliev O.L. (2012) Remodeling of nucleosome-dimer particles with yIsw2 promotes their association with ALL-1 SET domain in vitro. Biochemistry 49, 4354-4363.
- Krajewski W.A., Vassiliev O.L. (2011) Interaction of SET domains with histones and nucleic acid structures in active chromatin. Clinical Epigenetics 2, 17–25.
- Krajewski W.A., Vassiliev O.L. (2010) The S. cerevisiae Swi/Snf complex can catalyze formation of dimeric nucleosome structures in vitro. Biochemistry 49, 6531-6540.
- Krajewski W.A., Vassiliev O.L. (2010) Histone acetylation facilitates association of nucleosomes with SET domain of ALL-1 methyltransferase in vitro. Biochem Biophys Res Commun. 397, 112-116.
- Krajewski W.A., Reese J.C. (2010) SET domains of histone methyltransferases recognize ISWI-remodeled nucleosomal species. Mol Cell Biol. 30, 552-564.
Proteome regulation in invertebrates (sponges and lepidopterans)
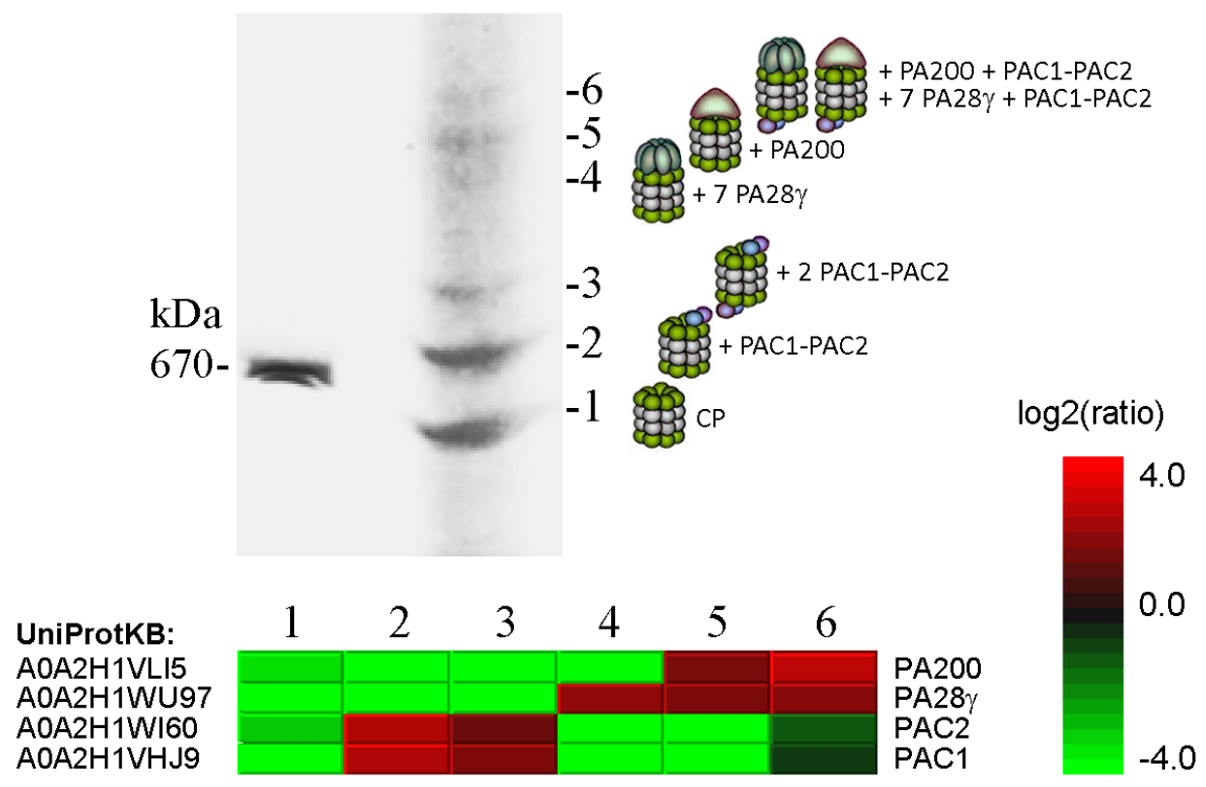
Fig. 3. Proteomic analysis of the six forms of 20S proteasome revealed after electrophoresis of Sf9 extracts in a native polyacrylamide gel. The upper panel shows a piece of gel stained with Coomassie blue and the bands numbered 1 to 6 taken for mass spectrometry (LC-MS/MS). Quantitative data on proteins significantly differing between the bands and the protein accession numbers in the UniProtKB database are shown in the heat map. Predicted composition of the six 20S proteasome complexes is shown to the right of gel. (From Kravchuk et al., 2019. BBA Proteins Proteom. 1867, 840-853).
Professor Victor S. Mikhailov and his colleagues investigate factors involved in the proteome regulation in lepidopteran insect Spodoptera frugiperda Sf9 cells at normal conditions and under infection with baculovirus (Autographa californica nucleopolyhedrovirus, AcMNPV). The virus infection was shown to induce a proteotoxic stress in infected cells that was accompanied by an accumulation of ubiquitinated proteins. The ubiquitin-proteasome system (UPS), chaperones of the HSP70/HSC70 family, and segregase VCP/p97 are involved in the proteome regulation in infected cells and are required for the viral genome replication and mature virus production. The proteome regulatory factors (proteasomes, chaperones, and segregase) of the lepidopteran insect cells are under investigation. The figure below illustrates fractionation by a native electrophoresis of 20S proteasome species in Sf9 cells.
Besides, the mechanisms of cell reaggregation in the sea cold-water sponge have recently begun to be studied. Regeneration is inherent in invertebrates and vertebrates. The ability to regenerate in the adult form is reduced in some higher vertebrates due to the development of adaptive immunity. Invertebrates, especially Sponges (Porifera), basal of Metazoa, have a significant potential for global regeneration. Sponges have no tissue, but their body is represented by different types of cells. Their distinctive feature is the ability to reaggregate cells after a complete dissociation of their body. Sponge cells are involved in constant movements, de- /transdifferentiations, and interactions with symbiotic microorganisms. Using the evolutionary approach, biochemical and molecular methods in the model system of sponge cell reaggregation, we reveal the similarities and differences in the processes of regeneration and repair in multicellular organisms.
Recent publications
- Kravchuk O.I., Lyupina Y.V., Erokhov P.A., Finoshin A.D., Adameyko K.I., Mishyna M.Yu, Moiseenko A.V., Sokolova O.S., Orlova O.V., Beljelarskaya S.N., Serebryakova M.V., Indeykina M.I., Bugrova A.E., Kononikhin A.S., Mikhailov V.S. Characterization of the 20S proteasome of the lepidopteran, Spodoptera frugiperda // Biochimica et Biophysica Acta (BBA) – Proteins and Proteomics. 2019. V. 1867. Is. 9. P. 840-853. DOI: 10.1016/j.bbapap.2019.06.010.
- Finoshin A.D., Adameyko K.I., Mikhailov K.V., Kravchuk O.I., Georgiev A.A., Gornostaev N.G. et al. (2020) Iron metabolic pathways in the processes of sponge plasticity. PLoS ONE 15(2): e0228722. https://doi.org/10.1371/journal.pone.0228722
- Adameyko K.I., Kravchuk O.I., Finoshin A.D., Mikhailov V.S., Gornostaev N.G., Lyupina Y.V., Bonchuk A.N., Georgiev A.A., Mikhailov K.V., Bacheva A.V., Indeykina M.I., Bugrova A.E., Gazizova G.R., Kozlova O.S., Gusev O.A., Shagimardanova E.I. Structure of neuroglobin from cold-water sponge Halisarca dujardinii // Molecular Biology. 2020. Т. 54. № 3. С. 416-420
Databases:
Transcriptomes: Halichondria panicea PRJNA594151 (NCBI), Halisarca dujardinii PRJNA594150 (NCBI)
Proteins: 38 UPS proteins (AZI95687.1 - AZI95650.1), 72 iron metabolism proteins QOI16596.1-QEH04755 (NCBI)