About the Functional Genomics Lab
Fields of Research
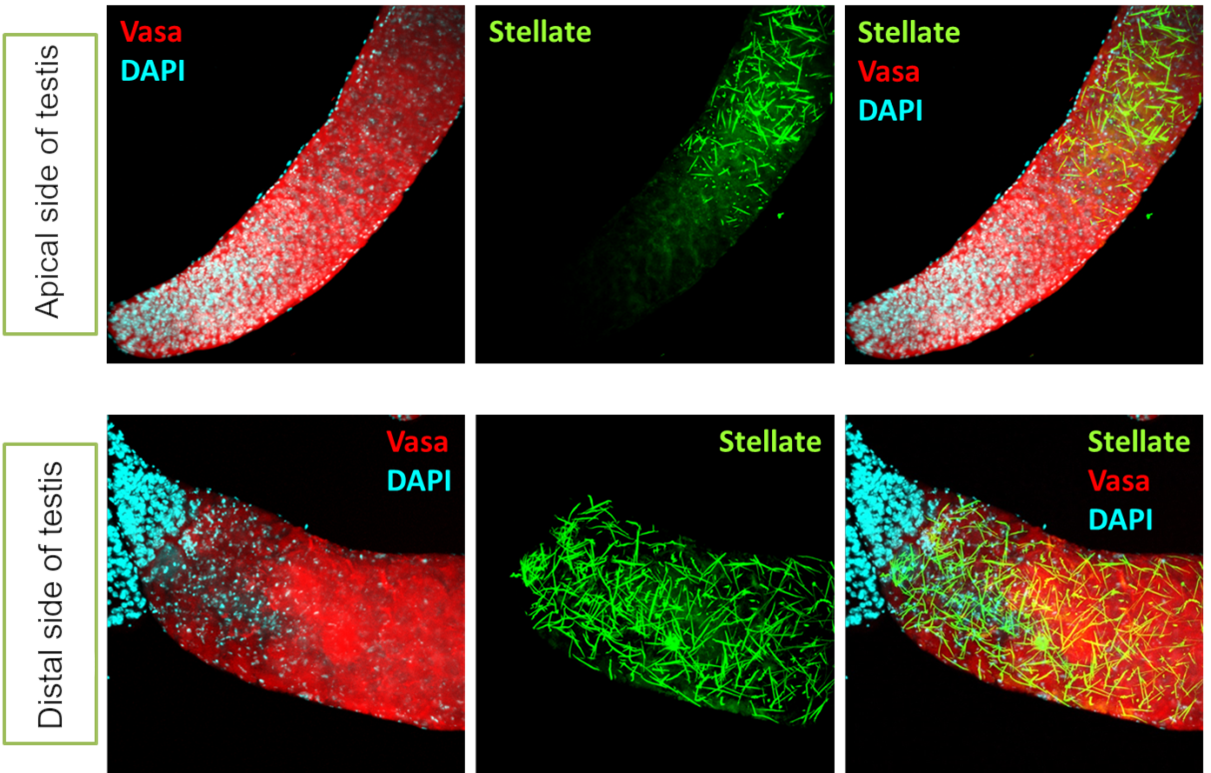
Fig. 1 Derepression of Stellate genes in the testes of interspecific hybrids obtained by crossing D. melanogaster and D. mauritiana.
Mechanisms of the piRNA pathway and its non-canonical functions
Small non-coding piRNAs (piwi-interacting RNA) of 24-32 nt in length are mostly expressed in germ cells, where their main function is to control mobile elements and ensure genome stability. Disruption of piRNA pathway activity leads to mobile elements activation, gametogenesis defects and sterility. piRNAs are produced from specific regions of the genome (piRNA clusters) and function in a complex with proteins of the PIWI subfamily of the ARGONAUTE family, providing target suppression at the post-transcriptional level or at the transcriptional level.
Previously, we have demonstrated important aspects of piRNA biogenesis and studied the functions of the piRNA pathway components - the Vasa and SpnE proteins (Kibanov et al., 2011; Ryazansky et al., 2016; Adashev et al., 2024). The piRNA pathway components evolve rapidly due to the need for the organism to adapt to the invasion of mobile elements. In addition to mobile elements, piRNAs can participate in the regulation of the protein-coding genes expression.
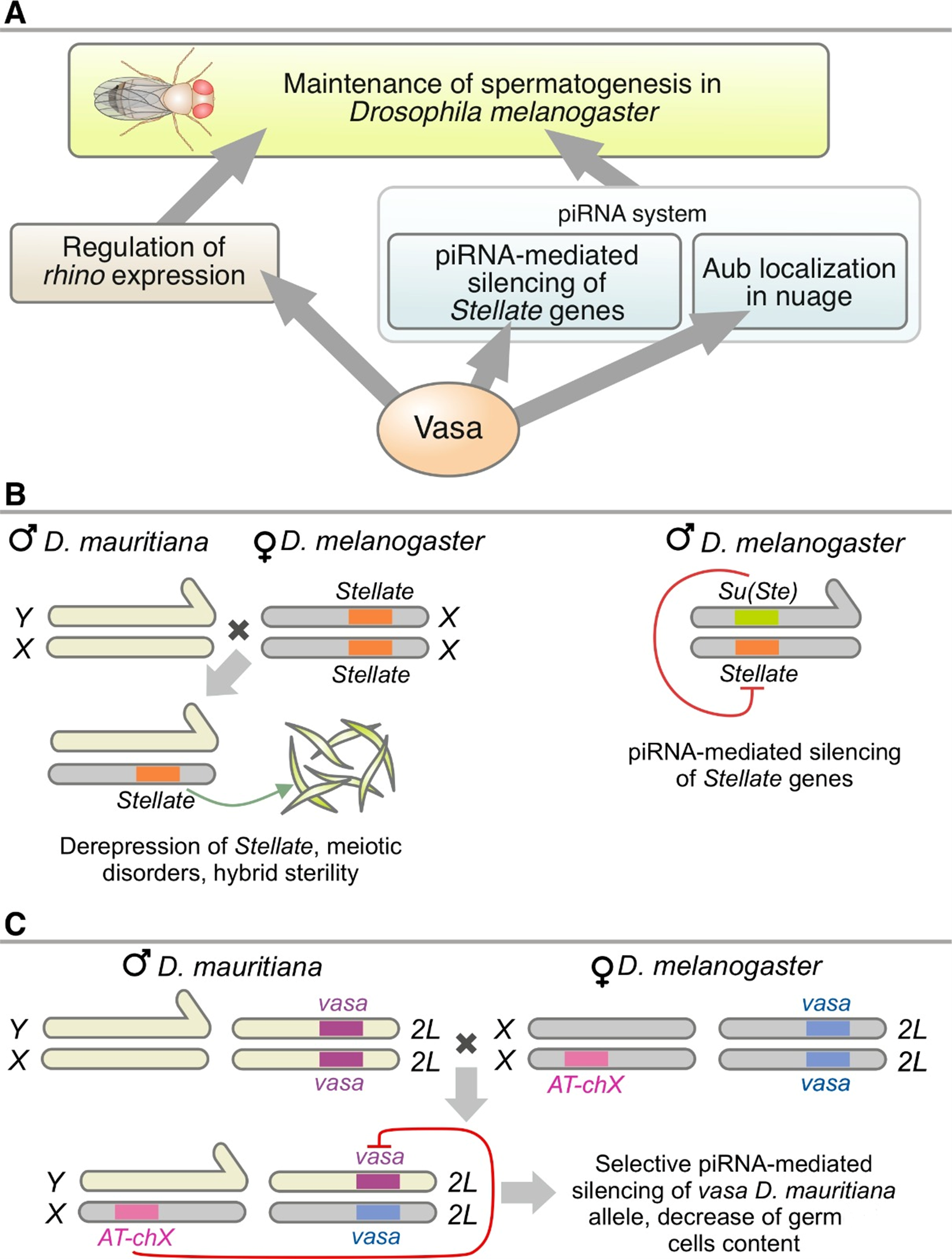
Fig. 2 Models of piRNA-dependent deregulation of Stellate and vasa genes in the testes of interspecific hybrids. A - Vasa functions in maintaining spermatogenesis in D. melanogaster. B – mechanism of Stellate derepression in testes of interspecific hybrids (left) and operation of the Stellate/Su(Ste) system in D. melanogaster males (right). С – mechanism of resulting vasa haploinsufficiency in males of interspecific hybrids.
Our studies have shown a role for the piRNA pathway in hybrid sterility formation through the non-canonical function of this pathway in regulating the protein-coding genes vasa and Stellate (Kotov et al., 2019; Adashev et al., 2021). In a collaboration with colleagues at Caltech, a sex-dependent dimorphism of transposon expression and piRNA pathway function was shown in D. melanogaster (Chen et al., 2021). Differences in the expression pattern of piRNA pathway components and mobile elements in the ovaries of hybrids between D. melanogaster and D. simulans have been demonstrated (Kotov et al., 2024) (Figs. 1, 2).
Thus, one of the directions of our research is to study the mechanism of piRNA silencing, its non-canonical functions in the regulation of protein-coding genes and the formation of hybrid sterility using classical and modern genetic, biochemical, molecular and bioinformatics approaches.
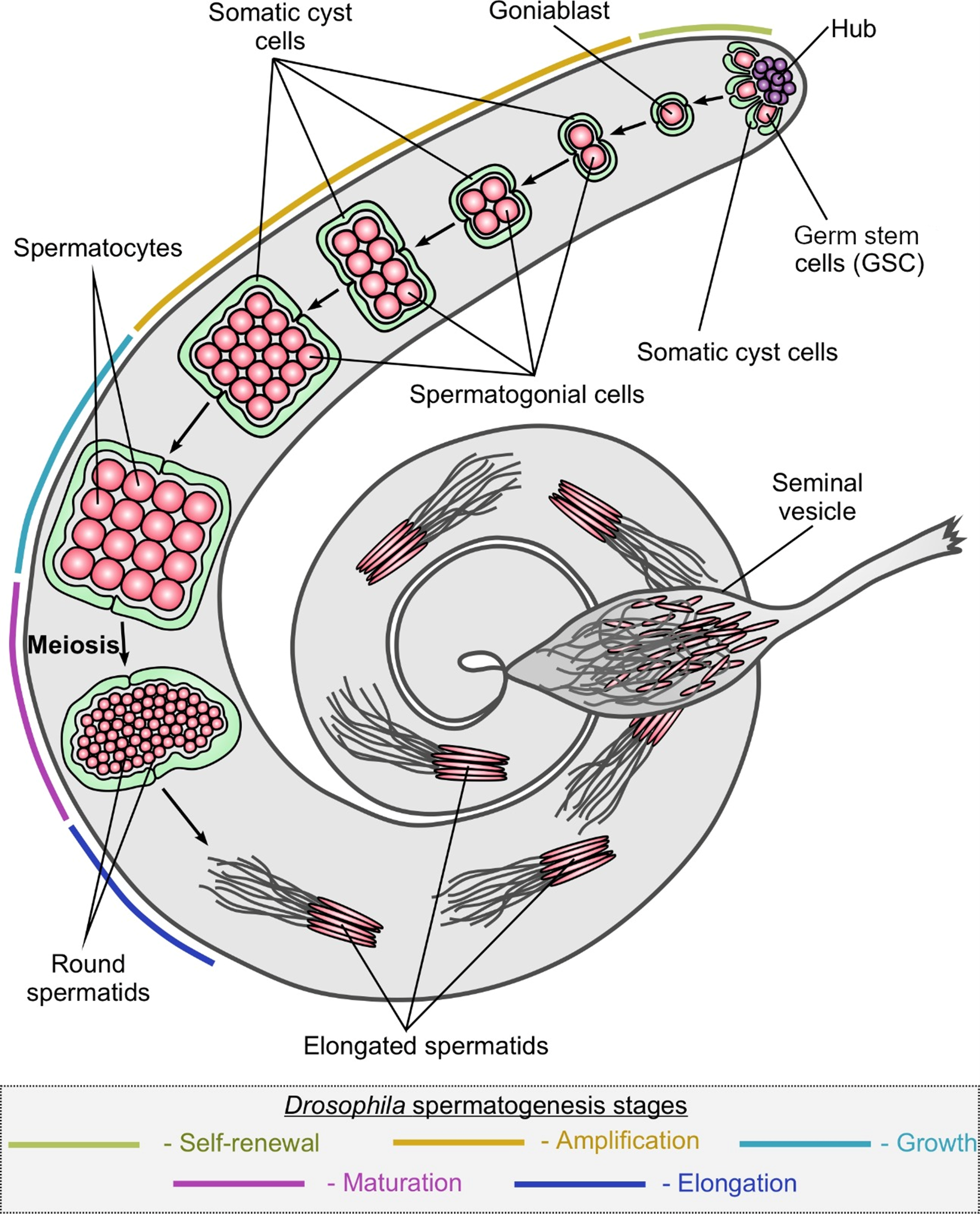
Mechanisms of germ stem cell maintenance and differentiation
Another focus of our laboratory is the study of molecular mechanisms underlying the regulation of germ cell maintenance and differentiation in the Drosophila model. The maintenance of germ stem cells (GSCs) is an important process that ensures reproductive function. HSCs are located in a specific microenvironment of supporting somatic cells, whose interaction maintains the balance of self-renewal and differentiation. This involves a fine regulation through various signals from the niche to stem cells, activity of transcription factors and metabolic processes in the body. We have previously shown essential functions of RNA helicase Belle (DDX3), an ortholog of human DDX3 (DBY), in the maintenance and differentiation of GSCs (Kotov et al., 2016; Kotov et al., 2020). We analyzed differential gene expression in early and mature somatic cells of Drosophila testis cysts using transcriptome libraries we obtained. An increased expression level of genes encoding components of a number of protein complexes was found at different stages of spermatogenesis. Differences in metabolic status between early and mature somatic cyst cells were found (Adashev et al., 2022). We have identified a novel mechanistic link between Vasa and Rhino, components of the piRNA pathway, in the regulatory network that mediates germ stem cell maintenance (Adashev et al., 2024) (Fig. 3).
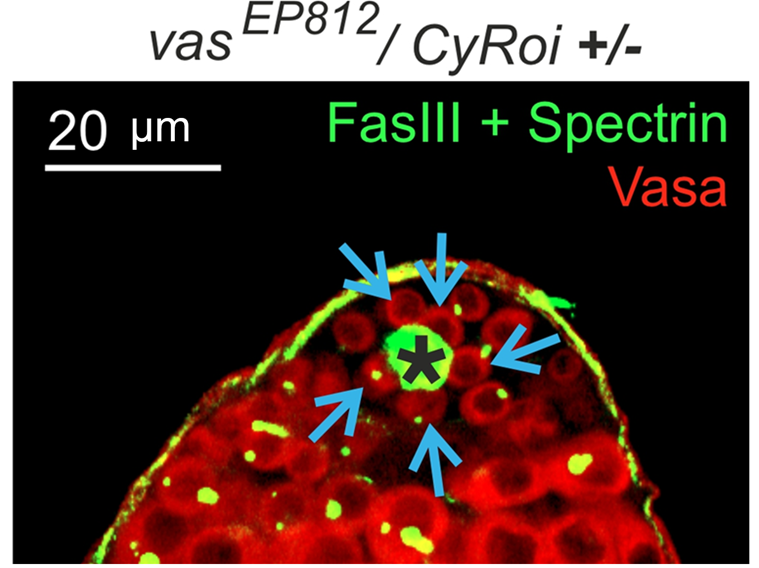
Fig. 3A Germ stem cells (blue arrows) in the Drosophila testis are located at the apical end around a structure of terminally differentiated somatic cells (hub, marked with *).
Members of Functional Genomics Lab

Alexei Kotov
Head of the Lab
PhD
Scientific interests: : mechanisms of stem cells maintenance and self-renewal, molecular features and non-canonical functions of the piRNA pathway, mobile elements, reproductive isolation, speciation and evolution, genome stability, bioinformatics, molecular biology and genetics

Ludmila Olenina
PhD, Leading Researcher
Scientific interests: regulation of mobile elements, piRNA pathway, transgenerational epigenetic inheritance, reproductive isolation, germline stem cells.

Aleksei Shatskikh
PhD, Senior Researcher
Scientific interests: mechanisms of gene expression regulation, non-coding RNA, chromatin, epigenetics.

Vladimir Adashev
PhD, Junior Researcher
Scientific interests: genetics of sex, epigenetic regulation of gene activity, developmental genetics, gametogenesis, piRNA silencing, interspecific hybrids, hybrid dysgenesis, evolutionary genetics.

Angelina Akishina
PhD, Junior Researcher
Scientific interests: developmental genetics, morphogenesis, transcription factors and transcription regulation, proliferation control

Elena Belkina
PhD, Junior Researcher
Scientific interests: evolutionary biology, reproductive isolation mechanisms, quantitative traits, molecular genetics, population genetics

Ekaterina Yakovleva
PhD, Junior Researcher
Scientific interests: mechanisms of adaptation to stressful environment, experiments on Drosophila melanogaster, evolution of life history traits, insect genomics, systems biology, bioinformatics, statistics and data visualization in R and Python

Sergei Bazylev
Senior laboratory assistant
Scientific interests: molecular biology, bioinformatics

Prokhor Proshakov
Senior laboratory assistant
Scientific interests: methods of genetic engineering, neurogenetics, behavioral genetics

Liliya A. Revyakina
Senior laboratory assistant
Scientific interests: genetics of aggressive behavior, population genetics, medical genetics, statistics

Ilia Kombarov
Senior laboratory assistant, PhD Student
Scientific interests: molecular genetics, short RNA, gametogenesis, genome stability, regulation of gene expression, Drosophila
Thesis title: The role of piRNA pathway components in the regulation of protein-coding genes and gametogenesis in Drosophila melanogaster

Elizaveta Davydova
Senior laboratory assistant, PhD Student
Scientific interests: population and evolutionary genetics, molecular sequences evolution rate, developmental genetics
Thesis title: The role of genomic duplications in the formation and functional specialization of new genes in Drosophila
Financial Support
- The laboratory is financed within the State assignment of the Institute of Developmental Biology of the Russian Academy of Sciences No. 0088-2024-0017.
- Some Lab members participate in the RSF project No. 25-28-00990 "Molecular and genetic components of aggressive behavior in Hadza and Dotoga children in the context of ontogenetic development" (headed by O.E. Lazebny).
Collaborations
- Institute of Molecular Genetics of the Russian Academy of Sciences (IMG RAS)
- Department of Molecular Genetics of the Federal State Budgetary Scientific Institution "VNIRO"
- Our laboratory collaborates with other laboratories of the IDB RAS
Selected publications
1. Adashev V.E., Kotov A.A., Bazylev S.S., Kombarov I.A., Olenkina O.M., Shatskikh A.S., Olenina L.V. Essential functions of RNA helicase Vasa in maintaining germline stem cells and piRNA-guided Stellate silencing in Drosophila spermatogenesis // Frontiers in Cell and Developmental Biology. 2024. Vol. 12. 1450227. DOI: 10.3389/fcell.2024.1450227.
2. Kotov A.A., Adashev V.E., Kombarov I.A., Bazylev S.S., Shatskikh A.S., Olenina L.V. Molecular Insights into Female Hybrid Sterility in Interspecific Crosses between Drosophila melanogaster and Drosophila simulans // International Journal of Molecular Sciences. 2024. Vol. 25. № 11. 5681. DOI: 10.3390/ijms25115681.
3. Sivunova D., Yakovleva E., Naimark E., Lysenkov S., Perfilieva K., Markov A. Adaptation of Drosophila melanogaster to high and low osmolarity promotes evolutionary change in the phenotypic plasticity of the larval anal organs // Biological Communications. 2024. № 3. P. 149-161. DOI: 0.21638/spbu03.2024.303.
4. Shatskikh A.S., Fefelova E.A., Klenov M.S. Functions of RNAi Pathways in Ribosomal RNA Regulation // Non-Coding RNA. 2024. Vol. 10. № 2. 19. DOI: 10.3390/ncrna10020019.
5. Yakovleva E., Danilova I., Maximova I., Shabaev A., Dmitrieva A., Belov A., Klyukina A., Perfilieva K., Bonch-Osmolovskaya E., Markov A. Salt concentration in substrate modulates the composition of bacterial and yeast microbiomes of Drosophila melanogaster // Microbiome Research Reports. 2024. Vol. 3. № 2. 19. DOI: 10.20517/mrr.2023.56.
6. Adashev V.E., Kotov A.A., Olenina L.V. RNA Helicase Vasa as a Multifunctional Conservative Regulator of Gametogenesis in Eukaryotes // Current Issues in Molecular Biology. 2023. Vol. 45. P. 5677-5705. DOI: 10.3390/cimb45070358.
7. Belkina E.G., Seleznev D.G., Sorokina S.Y., Kulikov A.M., Lazebny O.E. The Effect of Chromosomes on Courtship Behavior in Sibling Species of the Drosophila virilis Group // Insects. 2023. Vol. 14. № 7. P. 609. DOI: 10.3390/insects14070609.
8. Vorontsova J.E., Akishina A.A., Cherezov R.O., Simonova O.B. A new insight into the aryl hydrocarbon receptor/cytochrome 450 signaling pathway in MG63, HOS, SAOS2, and U2OS cell lines // Biochimie. 2023. Vol. 207. P. 102-112. DOI: 10.1016/j.biochi.2022.10.018.
9. Adashev V.E., Bazylev S.S., Potashnikova D.M., Godneeva B.K., Shatskikh A.S., Olenkina O.M., Olenina L.V., Kotov A.A. Comparative transcriptional analysis uncovers molecular processes in early and mature somatic cyst cells of Drosophila testes // European Journal of Cell Biology. 2022. Vol. 101. № 3. 151246. DOI: 10.1016/j.ejcb.2022.151246.
10. Chekunova A.I., Sorokina S.Y., Sivoplyas E.A., Bakhtoyarov G.N., Proshakov P.A., Fokin A.V., Melnikov A.I., Kulikov A.M. Episodes of Rapid Recovery of the Functional Activity of the ras85D Gene in the Evolutionary History of Phylogenetically Distant Drosophila Species // Frontiers in Genetics. 2022. Vol. 12. 807234. DOI: 10.3389/fgene.2021.807234.
11. Fefelova E.A., Pleshakova I.M., Mikhaleva E.A., Pirogov S.A., Poltorachenko V.A., Abramov Yu.A., Romashin D.D., Shatskikh A.S., Blokh R.S., Gvozdev V.A., Klenov M.S. Impaired function of rDNA transcription initiation machinery leads to derepression of ribosomal genes with insertions of R2 retrotransposon // Nucleic Acids Research. 2022. Vol. 50. № 2. P. 867-884. DOI: 10.1093/nar/gkab1276.
12. Chen P., Kotov A.A., Godneeva B.G., Bazylev S.S., Olenina L.V., Aravin A.A. piRNA-mediated gene regulation and adaptation to sex-specific transposon expression in Drosophila melanogaster male germline // Genes & Development. 2021. Vol. 35. № 1112. P. 914-935. DOI: 10.1101/gad.345041.120.
13. Belkina E.G., Lazebny O.E., Vedenina V.Y. The importance of acoustic signals in multimodal courtship behavior in Drosophila virilis, D. lummei and D. littoralis // Journal of Insect Behavior. 2021. Vol. 34. P. 280-295. DOI: 10.1007/s10905-021-09788-8.
14. Bazylev S.S., Adashev V.E., Shatskikh, A.S., Olenina L.V., Kotov A.A. Somatic Cyst Cells as a Microenvironment for the Maintenance and Differentiation of Germline Cells in Drosophila Spermatogenesis // Russian Journal of Developmental Biology. 2021. Vol. 52. P. 16–32. DOI: 10.1134/S1062360421010021.
15. Kotov A.A., Godneeva B.K., Olenkina O.M., Adashev V.E., Trostnikov M.V., Olenina L.V. The Drosophila RNA Helicase Belle (DDX3) Non-Autonomously Suppresses Germline Tumorigenesis via Regulation of a Specific mRNA Set // Cells. 2020. Vol. 9. № 3. 550. DOI: 10.3390/cells9030550.
16. Shatskikh A.S., Kotov A.A., Adashev V.E., Bazylev S.S., Olenina L.V. Functional Significance of Satellite DNAs: Insights From Drosophila // Frontiers in Cell and Developmental Biology. 2020. Vol. 8. 312. DOI: 10.3389/fcell.2020.00312.
17. Akishina A.A., Kuvaeva E.E., Vorontsova Y.E., Simonova O.B. NAP Family Histone Chaperones: Characterization and Role in Ontogenesis.// Russian Journal of Developmental Biology. 2020. Vol. 51. № 6. С. 343–355. DOI: 10.1134/S1062360420060028.
18. Kotov A.A., Adashev V.E., Godneeva B.K., Ninova M., Shatskikh A.S., Bazylev S.S., Aravin A.A., Olenina L.V. piRNA silencing contributes to interspecies hybrid sterility and reproductive isolation in Drosophila melanogaster // Nucleic Acids Research. 2019. Vol. 47. № 8. P. 4255-4271. DOI: 10.1093/nar/gkz130.
19. Akishina A.A., Vorontsova J.E., Cherezov R.O., Slezinger M.S., Simonova O.B., Kuzin B.A. NAP Family CG5017 Chaperone Pleiotropically Regulates Human AHR Target Genes Expression in Drosophila Testis // International Journal of Molecular Sciences. 2018. Vol. 20. № 1. 118. DOI: 10.3390/ijms20010118.
20. Belkina E.G., Naimark E.B., Gorshkova A.A., Markov A.V. Does adaptation to different diets result in assortative mating? Ambiguous results from experiments on Drosophila // Journal of Evolutionary Biology. 2018. Vol. 31. № 12. P. 1803-1814. DOI: 10.1111/jeb.13375.
21. Shatskikh A.S., Olenkina O.M., Solodovnikov A.A., Lavrov S.A. Regulated Gene Expression as a Tool for Analysis of Heterochromatin Position Effect in Drosophila // Biochemistry (Moscow). 2018. Vol. 83. P. 542–551. DOI: 10.1134/S0006297918050073.
22. Akishina A.A., Vorontsova J.E., Cherezov R.O., Mertsalov I.B., Zatsepina O.G., Slezinger M.S., Panin V.M., Petruk S., Enikolopov G.N., Mazo A., Simonova O.B., Kuzin B.A. Xenobiotic-induced activation of human aryl hydrocarbon receptor target genes in Drosophila is mediated by the epigenetic chromatin modifiers // Oncotarget. 2017. Vol. 8. № 61. P. 102934-102947. DOI: 10.18632/oncotarget.22173.
23. Abramov Y.A., Shatskikh A.S., Maksimenko O.G., Bonaccorsi S., Gvozdev V.A., Lavrov S.A. The Differences Between Cis- and Trans-Gene Inactivation Caused by Heterochromatin in Drosophila // Genetics. 2016. Vol. 202. № 1. P. 93-106. DOI: 10.1534/genetics.115.181693.
24. Yakovleva E.U., Naimark E.B., Markov A.V. Adaptation of Drosophila melanogaster to unfavorable growth medium affects lifespan and age-related fecundity // Biochemistry (Moscow). 2016. Т. 81. P. 1445-1460. DOI: 10.1134/S0006297916120063.
25. Kotov A.A., Olenkina O.M., Kibanov M.V., Olenina L.V. RNA helicase Belle (DDX3) is essential for male germline stem cell maintenance and division in Drosophila // Biochimica et Biophysica Acta. 2016. Vol. 1863. № 6 (Pt A). P. 1093-1105. DOI: 10.1016/j.bbamcr.2016.02.006.
26. Mitrofanov V.G., Chekunova A.I., Proshakov P.A., Barsukov M.I. Universal intracellular transducer Ras and its role in the development of Drosophila// Russian Journal of Developmental Biology. 2013. Vol. 44. P. 245–253. DOI: 10.1134/S1062360413040073.
27. Kibanov M.V., Egorova K.S., Ryazansky S.S., Sokolova O.A., Kotov A.A., Olenkina O.M., Stolyarenko A.D., Gvozdev V.A., Olenina L.V. A novel organelle, the piNG-body, in the nuage of Drosophila male germ cells is associated with piRNA-mediated gene silencing // Molecular Biology of the Cell. 2011. Vol. 22. № 18. P. 3410-3419. DOI: 10.1091/mbc.E11-02-0168.